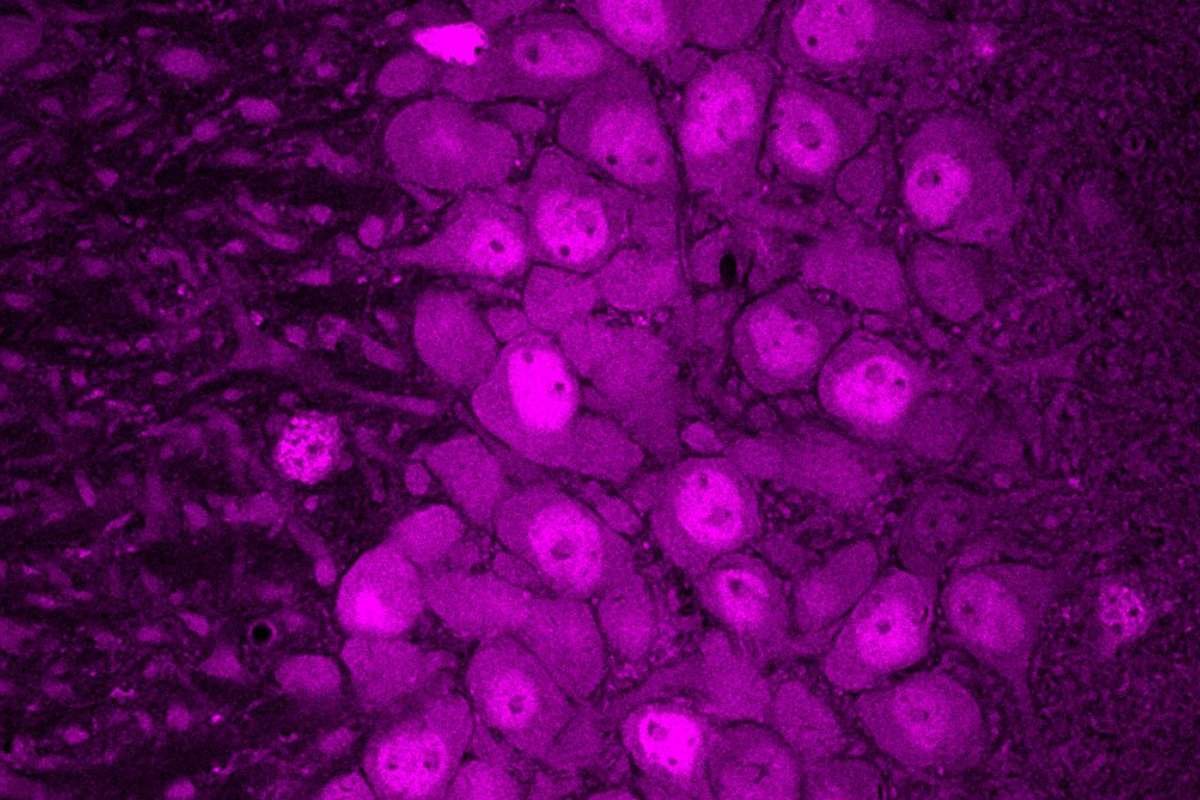
Researchers report a breakthrough in mapping how individual RNA isoforms translate into proteins in mouse brain cells, using a combined Ribo-STAMP and long-read single-cell sequencing approach to uncover cell-type-specific regulation.
Researchers Combine Technologies To Track Isoform Translation
A research team has mapped, for the first time at single-cell resolution, how distinct RNA isoforms are translated into proteins in the mouse brain Cells, addressing a longstanding challenge in neuroscience.
The study integrates Ribo-STAMP technology with MAS-ISO-seq long-read sequencing on PacBio’s Revio platform, enabling scientists to link translational activity directly to full-length RNA isoforms across thousands of individual cells.
By pairing RNA counts with measurements of cytidine-to-uridine, or C-to-U, editing events, researchers generated a quantitative metric known as EditsC. The measure reflects how actively each isoform engages with ribosomes while accounting for sequencing depth and genetic variation.
“This allows us to see which specific isoforms are being translated in each cell type, rather than inferring translation from bulk data,” the study’s lead investigator said in a statement. “It resolves a key blind spot in transcriptomics.”
The approach overcomes limitations of earlier short-read and bulk-tissue methods, which could not reliably assign translational signals to individual RNA variants in complex tissues such as the brain.
Findings Reveal Divergent Control Among Isoforms
Analysis shows that isoforms from the same gene often behave differently at the translational level, even when their RNA abundance appears similar.
While RNA expression levels between isoform pairs generally correlate positively across cell types, EditsC correlations frequently drop. More than one-third of isoform pairs show weak, zero, or negative translational correlations.
Many discordant pairs involve protein-coding isoforms and those retaining introns, a class of transcripts increasingly recognized for roles in mRNA stability and localization.
In neurons, isoforms with the highest EditsC values tend to have longer three-prime untranslated regions, or 3’ UTRs, more microRNA binding sites, and lower GC content. Researchers say these features may favor ribosome recruitment and translation.
“These structural elements appear to create a regulatory architecture that promotes translation in neurons,” a co-author said. “Non-neuronal cells such as astrocytes and oligodendrocytes show different patterns.”
The study also identifies neuron-specific ELAV-like proteins as positive regulators of translation. These RNA-binding proteins preferentially bind extended 3’ UTRs and interact with translation machinery.
Two isoforms of the synaptogenic gene Cadm3 illustrate the effect. Cadm3-202, which contains a longer 3’ UTR and enriched binding sites, shows higher EditsC values and greater translation than Cadm3-201.
Cell-Type Comparisons Highlight Translational Diversity
Comparisons across brain cell types reveal broad translational heterogeneity. Differences between oligodendrocytes and astrocytes produce the largest set of differentially translated transcripts.
Astrocytes show enrichment for transcripts with adenine-rich motifs in their UTRs, corresponding to binding sites for RNA-binding proteins linked to splicing and translation enhancement. Proteins such as Pabpc1, Pabpc4, and Sart3 are significantly enriched in astrocytes, the study reports.
In contrast, oligodendrocytes display enrichment of the RNA-binding protein Hnrnpr, suggesting distinct post-transcriptional regulatory networks.
The gene Rasal2 provides another example of cell-type-specific control. Its protein-coding isoform Rasal2-201 shows elevated translation in astrocytes, while the retained-intron isoform Rasal2-202 is preferentially translated in oligodendrocytes. Differences in RNA abundance do not fully explain the pattern.
“This points to translation-specific regulatory mechanisms, potentially mediated by intron retention,” the lead author said.
Researchers say the findings have implications for understanding synapse formation, plasticity, myelination, and neurological disease. By cataloging thousands of isoform-resolved translation events, the dataset offers a resource for future studies of the function of mouse brain cells and disorders.
The team adds that the method could extend beyond neuroscience to other tissues and disease models, enabling a detailed study of post-transcriptional regulation in health and disease.