A research team has successfully assembled the most complete and high-quality genome of Syntrichia caninervis, a moss species vital for maintaining ecological balance in the Gurbantunggut Desert. The findings, published in the Plant Biotechnology Journal, mark a significant advancement in plant genomics.
The study, led by Prof. Zhang Daoyuan from the Xinjiang Institute of Ecology and Geography (XIEG) under the Chinese Academy of Sciences, achieved a gapless genome assembly spanning 323.44 Mbp, with an N50 value of 24.41 Mbp and a single-base accuracy surpassing 99.999%. This makes it the most precise moss genome ever reported.
To facilitate this achievement, the researchers developed a monoclonal tissue culture system, successfully obtaining pure strains of Syntrichia caninervis. Utilizing advanced sequencing technologies such as PacBio and Oxford Nanopore, they also fully sequenced the moss’s endosymbiont, Paenibacillus sp., minimizing sequence contamination.
This Telomere-to-Telomere (T2T) genome surpasses previous attempts, particularly one based on the Mojave Desert population, by assembling over 30 Mbp of previously missing sequences, closing more than 20,000 chromosomal gaps, and identifying an additional 1,500 protein-coding genes. It also uncovered structural variations in sex chromosomes between ecotypes from the Gurbantunggut and Mojave Deserts.
Furthermore, researchers identified 677 transcription factor genes—135 more than before—and enhanced genome annotation. Transcriptomic mapping rates improved by 10%, and desiccation-related gene families, such as late embryogenesis abundant (LEA) genes, displayed increased tandem repeats.
The study also revealed that Syntrichia caninervis’s centromeric structure is dominated by Copia transposons, differing from angiosperms, offering new insights into land plant evolution. This comprehensive genome provides a strong foundation for future research in plant resilience and genetic adaptation.
News 2: Australian Scientists Develop Breakthrough Method to Clean Up Methylmercury Pollution

A team of Australian researchers has pioneered a new method to neutralize methylmercury, one of the world’s most hazardous pollutants. Their findings, published in Nature Communications on February 12, 2025, could pave the way for innovative approaches to protect both wildlife and human health.
Scientists from Macquarie University’s Applied BioSciences, CSIRO, Macquarie Medical School, and the ARC Centre of Excellence in Synthetic Biology successfully engineered fruit flies and zebrafish to transform toxic methylmercury into a far less harmful gas that dissipates into the air.
Dr. Kate Tepper, a synthetic biologist from Macquarie University and lead author of the study, expressed amazement at the discovery. “It still seems like magic to me that we can use synthetic biology to convert the most environmentally harmful form of mercury and evaporate it out of an animal,” she said.
Methylmercury, a byproduct of industrial activities like coal burning and illegal gold mining, poses serious risks due to its ability to accumulate in the environment and food chain. It can easily pass through the digestive system, blood-brain barrier, and placenta, leading to neurological and reproductive health issues.
The research team inserted bacterial gene variants into the DNA of fruit flies and zebrafish, enabling them to produce two enzymes that convert methylmercury into elemental mercury, which safely evaporates from their bodies.
“When we tested the modified animals, we found they retained less than half the mercury compared to unmodified ones, and most of it was in a far less toxic form,” Dr. Tepper explained.
While the study marks a significant step forward, researchers caution that extensive testing is needed to ensure both effectiveness and safety. Associate Professor Maselko emphasized the importance of regulatory oversight and built-in safety measures to prevent uncontrolled spread of the modified organisms in natural environments.
This breakthrough could lead to transformative strategies for mitigating mercury pollution and safeguarding ecosystems.
News 3: Scientists Develop Programmable CRISPR Delivery System for Targeted Gene Editing
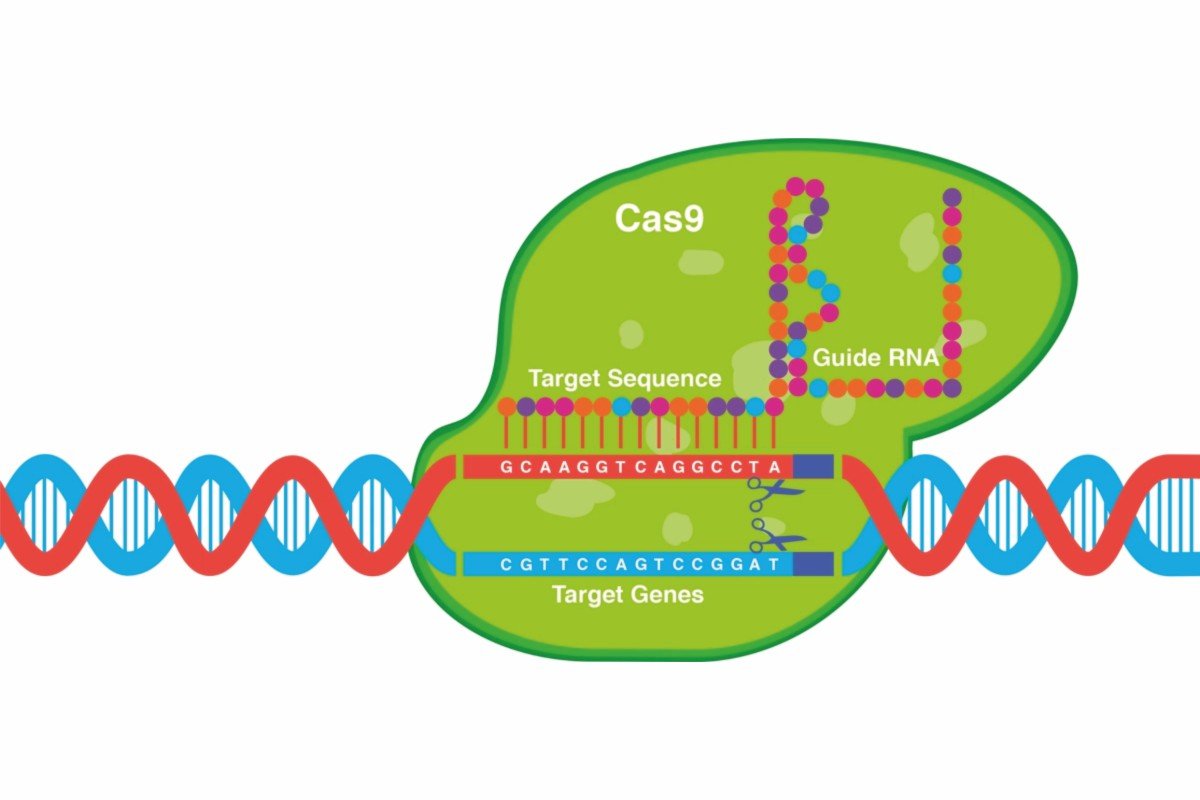
Researchers have developed a novel CRISPR-Cas9 delivery system called RIDE (Ribonucleoprotein delivery) that promises safer and more effective gene editing. RIDE utilizes virus-like particles (VLPs) to deliver CRISPR components, offering significant advantages over existing methods. The system can be readily reprogrammed to target specific cell types, including dendritic cells, T cells, and neurons, a crucial advancement for minimizing off-target effects and maximizing therapeutic benefit.
Current CRISPR delivery methods, like adeno-associated viruses (AAVs) and lipid nanoparticles (LNPs), often lack cell-type specificity, potentially leading to unintended gene editing and immune responses. RIDE addresses these limitations by leveraging the natural tropism of VLPs or engineering their surface proteins to target desired cells. The system assembles CRISPR ribonucleoproteins (RNPs) within the VLPs, ensuring efficient delivery and transient Cas9 expression, further reducing the risk of immunogenicity.
In preclinical studies, RIDE demonstrated comparable or superior gene editing efficiency compared to AAVs and LNPs. Researchers successfully used RIDE to treat mouse models of ocular neovascular disease and Huntington’s disease, achieving significant symptom improvement through cell-specific gene editing. Importantly, RIDE showed promising safety profiles in non-human primates, with minimal immune activation and no observable adverse effects. Furthermore, RIDE efficiently edited the huntingtin gene in neurons derived from Huntington’s disease patients’ induced pluripotent stem cells, demonstrating its potential for treating inherited neurological disorders.
The researchers also investigated the immune response to VLPs, finding that while some immune activation occurred, RIDE elicited a less pronounced anti-Cas9 antibody response compared to traditional lentiviral vectors. Single-cell RNA sequencing revealed changes in the immune microenvironment and cell state transitions, highlighting the importance of considering these factors in gene therapy.
RIDE’s programmable nature and favorable safety profile make it a promising platform for developing in vivo CRISPR therapeutics for a wide range of diseases. The ability to customize cell tropism opens up new avenues for targeted gene editing, paving the way for more precise and effective treatments.