Source-News-Medical
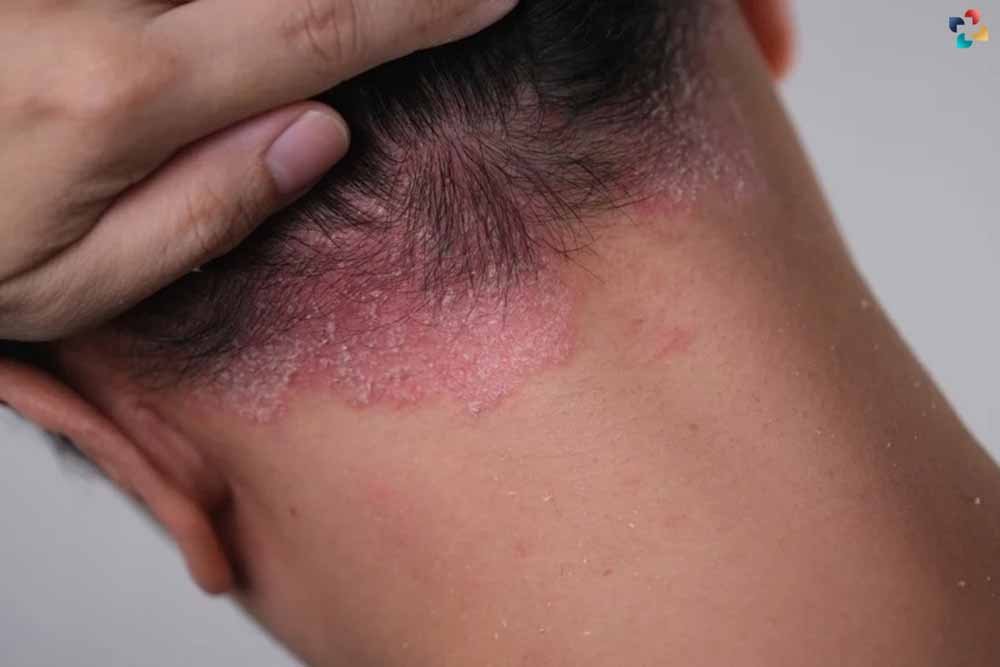
Psoriasis, a prevalent systemic inflammatory disorder affecting approximately 3% of the global population, extends beyond its visible symptoms, including its impact on the Skin Microbiome. Known for causing conditions like diabetes, psoriatic arthritis, and cardiovascular disease, it remains a significant health challenge due to its chronic nature and the lack of a definitive cure. The disease manifests in various clinical subtypes, influenced by factors ranging from the skin’s barrier function to genetic predisposition and environmental triggers. Central to recent research is the role of the skin microbiome—the diverse community of microorganisms residing on the skin—in the development and progression of psoriasis.
Insights from the Multi-Omics Study
A recent study conducted a comprehensive analysis to explore the intricate relationship between the skin microbiome and psoriasis severity. Led by researchers using data from the Microbes in Allergy and Autoimmunity Related to the Skin (MAARS) cohort, the study enrolled 116 individuals with plaque-type psoriasis alongside 102 healthy volunteers. Notably, participants with recent antibiotic use or undergoing specific treatments were excluded to isolate the impact of the disease on the microbiome.
Skin biopsies and microbiome samples were meticulously collected from active psoriatic lesions and adjacent non-lesional skin areas on the lower back. These samples were then subjected to advanced genomic analysis: DNA from microbiome samples underwent shotgun metagenomic sequencing, while RNA from biopsies was analyzed for gene expression patterns using weighted gene correlation network analysis (WGCNA).
Findings and Implications
The findings revealed distinct microbial and genetic signatures associated with psoriasis severity. The skin transcriptome of psoriatic lesions differed significantly from non-lesional skin, highlighting specific gene expression profiles linked to inflammation and immune response pathways. Notably, modules of genes identified through WGCNA showed correlations with the severity of psoriasis, as measured by the Psoriasis Area and Severity Index (PASI).
Moreover, the study identified microbial species, including Corynebacterium simulans, whose abundance correlated positively with the presence of immune cells like CD4 T cells and dendritic cells in psoriatic lesions. Conversely, Cutibacterium acnes, typically more abundant in healthy skin, showed reduced presence in psoriatic lesions. This dysbiosis in the skin microbiome was further underscored by distinct clusters of microbial gene families associated with different metabolic and inflammatory responses in psoriatic lesions.
In conclusion, the study not only deepened the understanding of host-microbe interactions in psoriasis but also hinted at potential therapeutic avenues. By targeting both the skin microbiome and immunological pathways, future treatments could aim to modulate disease severity more effectively. This dual approach underscores the complexity of psoriasis and the promising prospects for personalized medicine in managing this challenging condition.