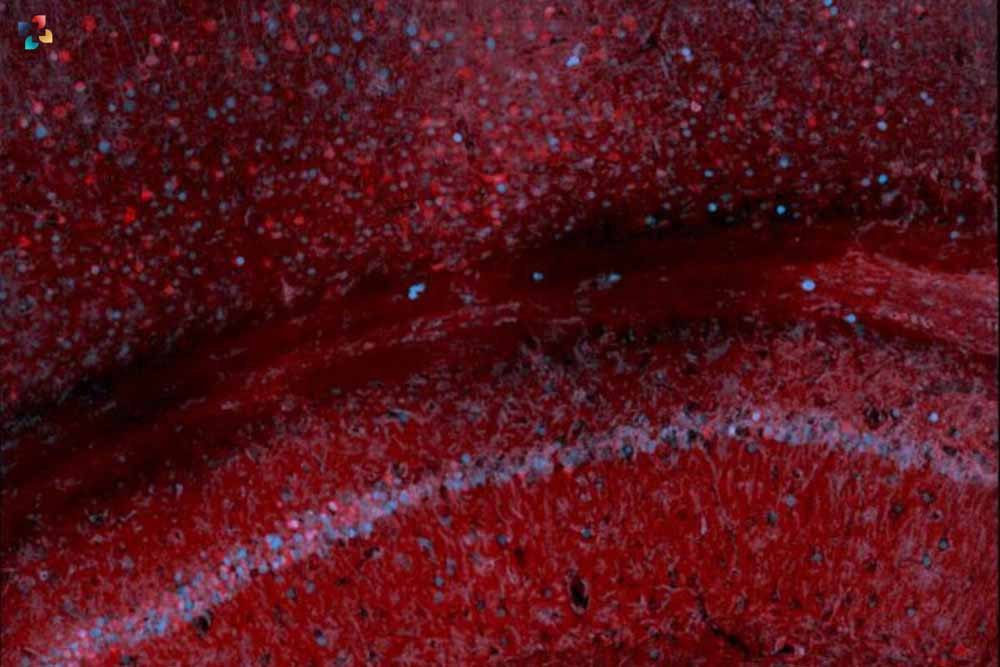
With its intricate network of specialised blood arteries forming an almost impenetrable barrier, the brain is the most protected organ in the body. This special anatomical configuration shields it from external attackers, but it also makes it challenging for researchers to investigate the expression of individual genes and how variations in gene expression can result in disease.
According to a paper published in Nature Biotechnology, researchers at Rice University have now created a noninvasive method of tracking the dynamics of gene expression in the brain, which will facilitate future research into neurological disorders, cognitive function, and brain development.
Released markers of activity (RMAs) are a novel class of molecules that have been created by Rice bioengineer Jerzy Szablowski and colleagues. These molecules can be used as a simple blood test to detect gene expression in the brain.
“Typically, if you wanted to look at gene expression in the brain, you would have to wait to do a post-mortem analysis. There are some more modern neuroimaging techniques that can do this, but they lack sensitivity and specificity to track changes in specific cell types.
With the RMA platform, we can introduce a synthetic gene expression reporter to the brain, which produces a protein that can pass through the blood-brain barrier. We can then measure changes in expression for a gene of interest with a simple blood test.”
Jerzy Szablowski, assistant professor of bioengineering at Rice’s George R. Brown School of Engineering
Szablowski saw that the brain would rapidly clear injections of antibody treatment, thus he initially thought of the prospect of a synthetic gene expression reporter.
“Whenever these injections were done, the antibodies would just disappear ⎯ they wouldn’t hang around long enough in the brain for an effective therapy,” he said. However, we believed that the shortcomings of antibody treatments could be used to our benefit. What would happen if we connected the portion of the antibody that caused this escape to a protein that was simple to identify? Then, we could observe the location, timing, and degree of a certain gene’s expression in the brain.”
Brain Secrets in a Blood Drop: Dynamics Of Gene Expression In The Brain
Previous studies had already established that the neonatal fragment crystallizable receptor (FcRn), a gene involved in regulating the quantity of antibodies throughout the body, is how antibodies penetrate the blood-brain barrier. To take advantage of this biological escape hatch, Szablowski and colleagues used advanced bioengineering techniques to connect the portion of the antibody that facilitates passage through the blood-brain barrier to a common reporter protein. The researchers were subsequently able to observe that expression mirrored in the mouse’s blood when they linked the RMAs to a particular gene and expressed that gene in the mouse’s brain.
“This method is very sensitive and can track changes in specific cells,” Szablowski stated. When this protein was produced in about 1% of the brain, blood levels of it increased up to 100,000 times over baseline. With only a blood test, we could precisely monitor the expression of this one protein.”
For the time being, RMAs are crucial research instruments in Szablowski’s opinion, helping scientists better track gene expression in the brain. He mentioned that the RMA platform might be utilised to examine the duration of time that innovative gene treatments remain in the brain.
Because the RMA platform is noninvasive, we could monitor these novel therapies over time and track them with simply a blood test,” he said. But RMAs can also be used to investigate the relationship between disease and gene expression. We will be able to learn more about the causes of disease and how it alters gene expression in the brain if we can monitor various changes in gene expression. This may offer fresh insights for the creation of medications or possibly for the prevention of neurological illnesses altogether.”
The David and Lucile Packard Foundation (2021-73005) and the National Institutes of Health (R21EB033059, DP2GM140923, R00DA043609, F31NS125927) supported the research.